Phys.org October 3, 2022
Researchers at UC Riverside developed a model to explore the mechanisms of RNA condensation by structural proteins, protein oligomerization and cellular membrane–protein interactions that control the budding process and the ultimate virus structure. Using molecular dynamics simulations, they deciphered how the positively charged N proteins interact and condense the very long genomic RNA resulting in its packaging by a lipid envelope decorated with structural proteins inside a host cell. Considering the length of RNA and the size of the virus they found that the intrinsic curvature of M proteins is essential for virus budding. While most current research has focused on the S protein, which is responsible for viral entry, and it has been motivated by the need to develop efficacious vaccines, the development of resistance through mutations in this crucial protein makes it essential to elucidate the details of the viral life cycle to identify other drug targets for future therapy. According to the researchers their simulations will provide insight into the viral life cycle through the assembly of viral particles de novo and potentially identify therapeutic targets for future drug development…read more. Open Access TECHNICAL ARTICLE
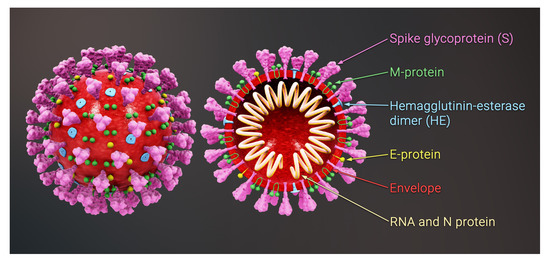
Schematic view of the structure of a coronavirus. …Credit: Viruses 2022, 14(10), 2089